News Express: UM collaborates with UK and US research teams to develop innovative computational model for electrophysiology experiments
新聞快訊:澳大與英美科研團隊開發創新電生理模型
膜片鉗實驗的新電腦模型揭示了被忽視的實驗偏移和延遲
The new computational model of the patch-clamp experiment reveals shifts and delays that were previously overlooked
澳大與英美科研團隊開發創新電生理模型
澳門大學健康科學學院助理教授李俊樂與英國諾丁漢大學、美國康奈爾大學的研究團隊合作,研發出新的膜片鉗實驗電腦模型,能夠更好理解電訊號在神經細胞、心臟細胞等細胞中的運作方式,揭示過去無法檢測的細胞電功能細節。該項新技術為電生理學研究領域開闢了新方向,有望為疾病診斷和藥物安全性檢測帶來新見解。相關研究已發表於國際頂尖期刊《先進科學》(Advanced Science)。
電生理學是一門研究細胞如何利用電訊號維持功能和進行溝通的科學,尤其聚焦於神經細胞和心臟肌肉細胞。這些電訊號對人體至關重要,關係到思考、活動和維持心臟跳動,是生命的“電語言”。膜片鉗實驗是該領域最重要的工具之一,能夠精確地測量單一細胞中微小的電變化。它的部分功能是測量和控制細胞的膜電位,同時測量流過細胞膜的離子電流,對於理解細胞如何產生電脈衝至關重要。
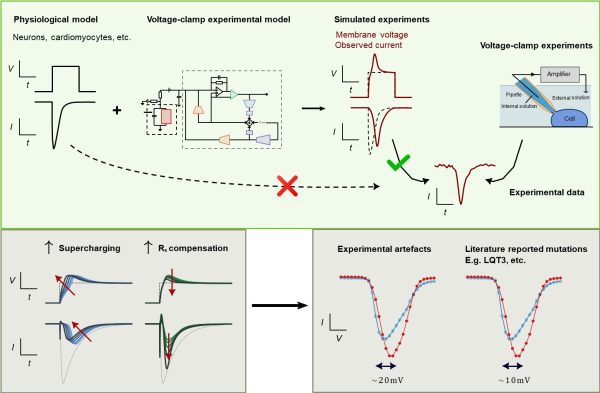
儘管膜片鉗實驗在細胞電生理學已有長期的研究基礎,但其電訊號難以準確地被測量及解讀。許多此類數據,包括與安全性相關的關鍵資訊,會受到膜片鉗實驗誤差的影響而導致被誤讀。為應對這一挑戰,研究團隊開發了一種創新的電腦模型,可有效識別和糾正膜片鉗實驗中的誤差。該新型電腦模型模擬了實驗工具和條件如何影響記錄資料,包括放大器的設定方式及細胞本身的生物特性。它整合了多項技術參數,如細胞電容、電阻與電壓偏移等,並已在多種實驗場景中進行測試,以及涵蓋不同類型的細胞和實驗設置,包括人類幹細胞衍生的心肌細胞和轉染腎細胞。
當該模型應用於在心肌細胞功能中發揮關鍵作用的心臟鈉電壓門控電流時,研究團隊通過關聯觀察到的電流與模擬的膜電壓起來,成功解決了實驗誤差問題,從而解釋了記錄電流中常見的偏移和延遲,這些現象在過去一直難以被準確理解。該研究還表明,將包含實驗誤差的電流電壓數據進行平均算法可解釋以往疾病相關基因突變或藥物作用的偏差,凸顯了使用該研究的電腦模型進行準確數據解釋的重要性。
透過提高這些膜片鉗實驗記錄的精確度,該研究能夠揭示細胞的真實電生理行為,挖掘出過去因技術限制而未能觀察到的重要訊息,使電生理學研究能夠在比以往更廣泛的實驗條件下擬合並恢復離子電流特性的精確估計值。該突破不僅提高了相關研究的可靠性,也有助於深化對心律不整、神經系統疾病以及其他與細胞電活動相關疾病或藥物作用的理解。
該研究的共同第一作者及通訊作者為李俊樂。美國康奈爾大學前研究員Alexander Clark參與了實驗工作,英國諾丁漢大學教授Gary Mirams協助設計了計算模型;其他團隊成員亦在研究的不同環節提供支援。該項目由澳門特別行政區科學技術發展基金(檔案編號:0155/2023/RIA3及0048/2022/A)和澳門大學(檔案編號:SRG2024-00014-FHS)資助。全文可瀏覽:https://advanced.onlinelibrary.wiley.com/doi/full/10.1002/advs.202500691。
欲瀏覽官網版可登入以下連結:
https://www.um.edu.mo/zh-hant/news-and-press-releases/campus-news/detail/61634/
UM collaborates with UK and US research teams to develop innovative computational model for electrophysiology experiments
Lei Chon Lok, assistant professor in the Faculty of Health Sciences (FHS) at the University of Macau (UM), in collaboration with research teams from the University of Nottingham and Cornell University, has developed a new computational model for patch-clamp experiments. This model enhances understanding of how electrical signals work in cells, such as nerve and heart muscle cells, by revealing previously undetectable details of how they function. The new technique opens up new avenues for electrophysiology research and provides fresh insights into disease diagnosis and drug safety testing. The research has been published in the top international journal Advanced Science.
Electrophysiology is the study of how cells, especially nerve and heart muscle cells, use electrical signals to function and communicate. These signals are vital to the human body, underpinning thought, movement, and the heartbeat. In other words, they are the ‘electrical language’ of life. One of the most important tools in this field is the patch-clamp experiment, which enables researchers to measure tiny electrical changes in individual cells with remarkable precision. This experiment measures and controls the membrane potential of a cell while simultaneously measuring the ionic currents flowing across the cell membrane, which is crucial for understanding how cells generate electrical impulses.
Although the patch clamp experiment has long been a cornerstone of cellular electrophysiology, scientists have often struggled to accurately measure and interpret electrical signals recorded from cells. Many of these recordings, including information crucial for safety considerations, can be misinterpreted due to subtle experimental distortions, also known as artefacts. To address this challenge, the research team has developed an innovative computational model that can effectively identify and correct these artefacts in patch-clamp experiments. The model simulates the influence of experimental tools and conditions, including amplifier settings and cell properties, on the recorded data. It integrates various technical parameters, such as capacitance, resistance, and voltage offset, and has been tested in a series of experiments involving different cell types and setups, including human stem cell-derived heart muscle cells and genetically modified kidney cells.
When the team applied the model to the cardiac fast sodium current, a key factor in heart muscle function, they successfully resolved experimental artefacts by linking observed current recordings with simulated membrane voltage. This explains the shifts and delays in recorded currents that have previously puzzled researchers. The study also showed that typical current-voltage data artefacts can explain biases that had previously been attributed to disease-related genetic mutations or drug effects, highlighting the importance of using the new computational model for accurate data interpretation.
Improving the precision of these patch-clamp recordings enables the computational model to reveal the true electrical behaviour of cells, providing access to important information that was previously obscured by technical limitations. This enables electrophysiological research to make fitting and recover precise estimates of ionic current properties under a wider range of experimental conditions. This breakthrough not only enhances the reliability of related research but also contributes to a deeper understanding of heart rhythm disorders, neurological disorders, and other diseases related to cellular electrical activity, as well as drug effects.
Prof Lei is the co-first and corresponding author of the study. Alexander Clark, a former researcher at Cornell University, contributed to the experimental work, and Gary Mirams, professor at the University of Nottingham, assisted in the design of the computational model. Other team members also provided support at different stages of the research. The project was funded by the Science and Technology Development Fund of the Macao SAR (File No.: 0155/2023/RIA3 and 0048/2022/A), and the University of Macau (File No.: SRG2024-00014-FHS). The full version of the research article is available at: https://advanced.onlinelibrary.wiley.com/doi/full/10.1002/advs.202500691.
To read the news on UM’s official website, please visit the following link:
https://www.um.edu.mo/news-and-press-releases/campus-news/detail/61634/